Table of Contents
How to... visualize min G* and P using GNUPlot
Compatibility
MatCalc version: -
Database: -
Author: Georg Stechauner
Created: 2013-03-06
Revisions: -
GNUPlot version: 4.6
Objectives
This second part shows a way to process the data calculated by MatCalc and how to create easily interpretable plots. The experienced user is free to use any program of his choice, which can process large amounts of data and plot them in 3D (e.g. Origin, MatLab, …). In order to provide a solution for everyone, an approach using GNUPlot (free, opensource software) is shown in this example.
Related documents
- http://www.gnuplot.info/ - GNUPlot official Homepage. Download software there, also FAQ and Demos.
Complementary files
Click here to view the GNUPlot script for this HowTo manual.
Main document
Export data to file
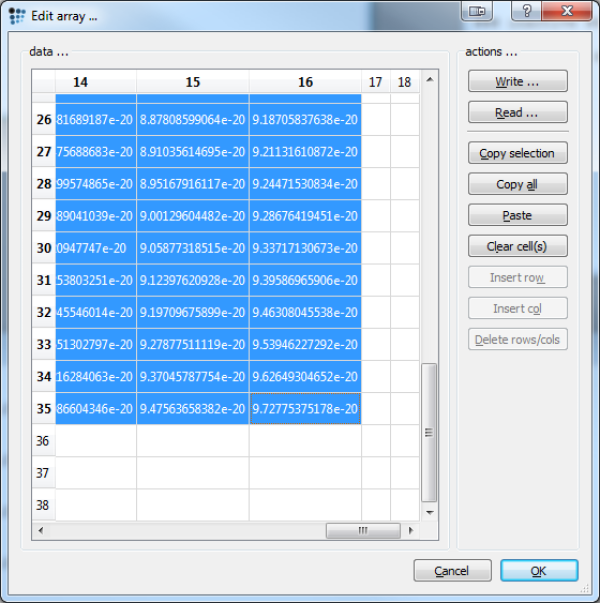
To create a 3-dimensional G*, respectively P plot, we need to extract the relevant data from MatCalc. Open the desired array (either G* or normalized P) and mark all data, without the axes (values for X and Y). Then press: Copy selection. Alternatively open the 'array_without_xy' array and press Copy all.
Next step, paste the values into an editor. Windows users can easily open the command line (Shortcut: WIN+R) and type 'cmd'.
The values in the editor should be separated with Tab-stops and have '.' as commas. This is important, because GNUPlot will not process ',' as a comma (This is especially important, if you copy the data to Excel and it changes the comma!).
Save the file as *.txt and name it anyway you want.
Note: As this file has no indication of X- and Y-axis (or rather column and row) description, either name the file properly, or create a second file which describes the variables. This will be needed later on in the script, when we label the axes.
Plot with GNUPlot
Executing the GNUPlot program file will open a command line program. The newest version 4.6 supplies some GUI which can help, if you are lost. The following procedure however is done by scripting only, without the GUI.
Start by pointing to the correct directory, so GNUPlot knows where the data-files are. Write the following command, or press 'ChDir' in the GUI:
cd 'C:\datafiles\VCN\'
Note: It is very important to always set file names and directories between quotation marks '…'. An easy way to do this, is to copy the filename/directory location in Explorer, and then use Right-click - Paste to enter it into GNUPlot.
After pointing to the correct directory we define axes and boundaries, and name everything. The hashtag # is used in GNUPlot as a comment, it is not necessary to type it.
set ticslevel 0.1 # Distance from data points to base plane
set zlabel 'min G*' # Label of Z-Axis
set ylabel 'wt-% V' # Label of Y-Axis
set xlabel 'wt-% N' # Label of X-Axis
set format z '%2.1e' # Display values on z axis as floating point with subsequent e
set cntrparam levels incr 1e-20,2e-20,1e-18 # Settings for contour lines.
# 1. Startvalue of contour lines, 2. Stepwidth between the
# contour lines - decrease this value, to increase amount of lines.
# 3. Finalvalue of contour lines.
set contours # Show contour lines
set style data lines # Connect data points with lines
set hidden3d # Hide points and lines which are hidden
set key outside # Show legend outside the plot
Attention: Set your own values, according to the results. Always double check the labeling: Z-Axis are the results (e.g. G* and P), X and Y are the variables which were used in the loops. Set the range of contourparameters in a way to capture all the important values! The values above will work with the G* file, created from part I.
splot 'VCN-gstar.txt' matrix # splot creates a 3D plot, matrix points to a matrix
# style data layout.
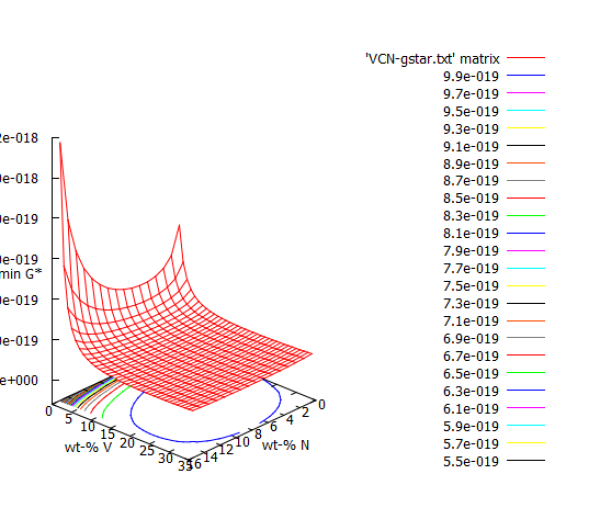
Click into the plot, and 'rotate' the picture a little bit. This will force the 'wxg graphic output terminal' (which is responsible for plotting data to your screen) to refresh. You can also click the 'Replot' button in the GUI bar on top of the output window. This has to done, whenever you change the size of the output window. If nothing crashed and no power shortages occured, you should have plot similar to this:
But what does this tell us? Why are the axes numbered from 0 to 35? The next section will handle these topics and refine the results.
Refine the plot
The problems which we have to solve in this section are:
- Too many, unnecessary contour lines, but…
- … not enough in the relevant area
- Wrong values on axes
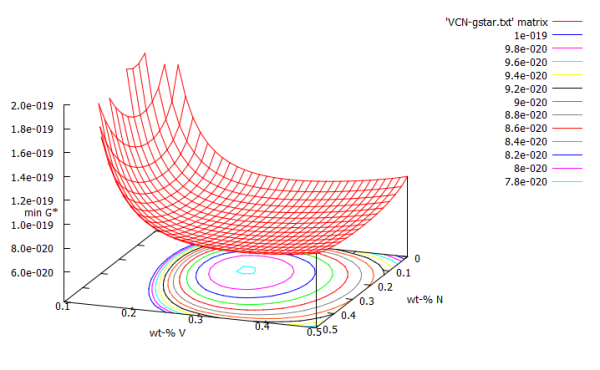
The first problem we formulated has to be approached by two ways: the first one is, to rescale the Z-axis, to the most relevant section, which lies between 5e-20 and 2e-19.
set zrange [5e-20:2e-19] replot
This code snippet rescales the z-range and automatically replots the picture. Now we need to adapt the contour lines:
set cntrparam levels incr 1e-20,2e-21,1e-19 replot
This leads to a high density of contours in the relevant area.
As a next step, we need to change X- and Y-axis to correct values. Due to the fact that we plotted a matrix, GNUPlot uses the Matrix-position of the data as X- and Y-values. As we use a 16×35 matrix, X-axis goes up to 16, and Y-axis to 35. It is the easiest way to manually set axes to correct values. Open the MatCalc script therefore, and open the G* array. Now you need 4-5 positions and name them:
set xtics ('0' 0, '0.1' 3, '0.2' 6, '0.3' 9, '0.4' 12, '0.5' 15)
set ytics ('0.1' 0, '0.2' 9, '0.3' 18, '0.4' 27, '0.5' 34)
replot
The command 'set xtics' lets the user manually set numbers on the axes. The digits between the apostrophes are the label, the following digit the position.
The final version of our min G* plot should look like this.
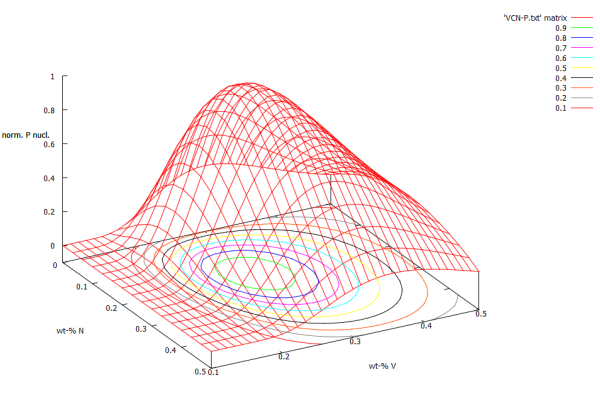
However, this is only a plot of energy, and does not represent the nucleation event anywhere as good as a normalized P-array plot. Use the previously gained knowledge, and create the normalized P-array plot now!
Plot of normalized nucleation probability
Without further ado, here are the code lines, to produce the corresponding graph:
cd 'C:\datafiles\VCN\'
set ticslevel 0.1 # Distance from data points to base plane
set zlabel 'norm. P nucl.' # Label of Z-Axis
set ylabel 'wt-% V' # Label of Y-Axis
set xlabel 'wt-% N' # Label of X-Axis
set cntrparam levels incr 0.1,0.1,1 # Settings for contour lines.
# 1. Startvalue of contour lines, 2. Stepwidth between the
# contour lines - decrease this value, to increase amount of lines.
# 3. Finalvalue of contour lines.
set contours # Show contour lines
set style data lines # Connect data points with lines
set key outside # Show legend outside the plot
set xtics ('0' 0, '0.1' 3, '0.2' 6, '0.3' 9, '0.4' 12, '0.5' 15)
set ytics ('0.1' 0, '0.2' 9, '0.3' 18, '0.4' 27, '0.5' 34)
splot 'VCN-P.txt' matrix # splot creates a 3D plot, matrix points to a matrix
# style data layout.
The careful reader has maybe noticed some missing commands. These are 'set hidden3d'… we are not using this command here, as the resulting mesh would totally cover the contour lines. Further we don't have to set a specific format on z axis.
Note: It is possible to create directly *.jpgs or *.pngs from GNUPlot. However, this is neither trivial nor error-resistant. The easiest way to get images from the plots is to use either print-screen or a snipping tool of your choice,
Additional commands
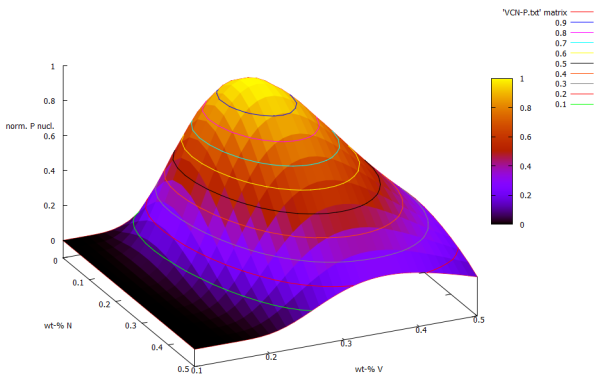
There are some (actually many) commands which can possibly enhance the picture. The following one 'set pm3d' will color-code numerical values directly to the surface. Use this command either with or without 'set hidden3d'. By using 'set contour surface' you can directly see the lines, even while using hidden3d
set pm3d set hidden3d set contour surface
Consecutive articles
For detailed commands and more information on GNUPlot, please visit their Homepage on demo files: http://www.gnuplot.info/demo/
Other tutorials can be found on: http://www.gnuplot.info/help.html